Ensembl Variation - Schema documentation
This document gives a high-level description of the tables that make up the Ensembl variation schema. Tables are listed by alphabetical order, and the purpose of each table is explained. It is intended to allow people to familiarise themselves with the schema when encountering it for the first time, or when they need to use some tables that they've not used before.
This document refers to version 104 of the Ensembl variation schema.
The variation database schema diagram (PDF format) is available here:
List of the tables:
Attributes tables
These tables define the variation attributes data.
Defines various attributes used elsewhere in the database
Example:
See below the query to display a subset of the attrib entries:
SELECT * FROM attrib WHERE attrib_type_id IN (469,470,471) ORDER BY attrib_id LIMIT 21;
| See also: |
List of species with populated data:
|
Groups related attributes together
| See also: |
List of species with populated data:
|
Defines the set of possible attribute types used in the attrib table
Example:
See below the command to display a subset of the attrib_type entries:
SELECT * FROM attrib_type WHERE attrib_type_id > 468 LIMIT 10;
| See also: |
List of species with populated data:
|
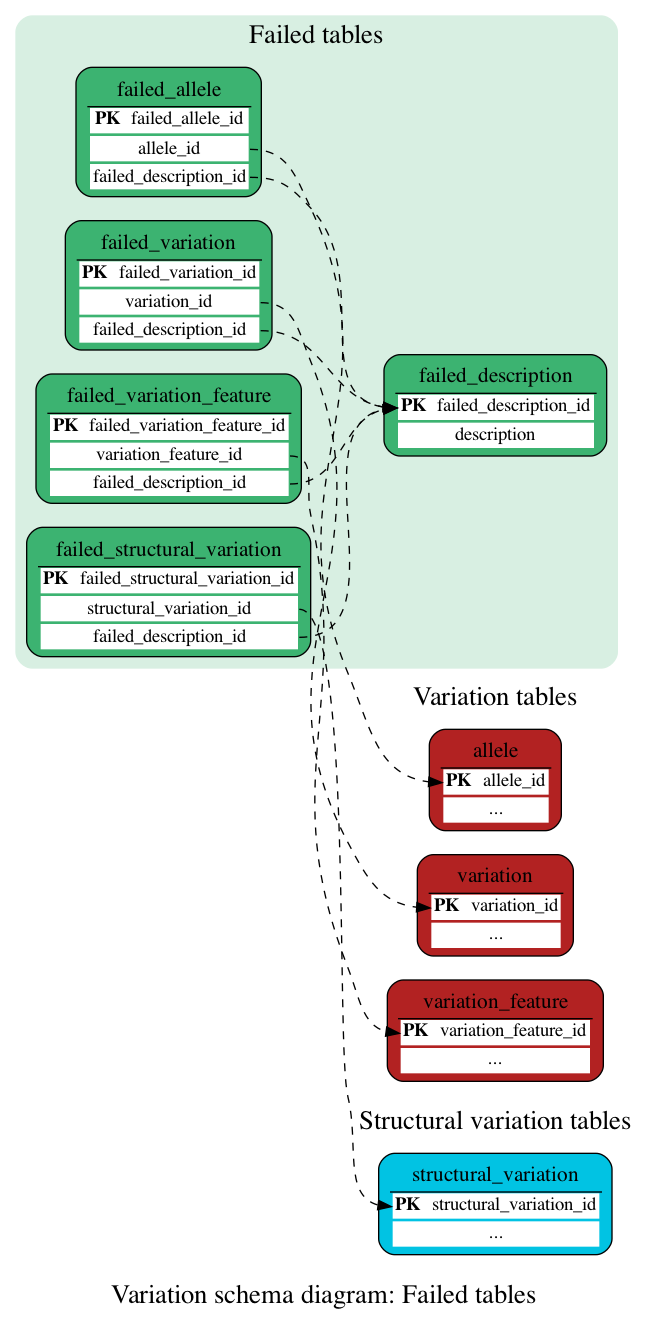
Failed tables
These tables define the list of variants/alleles flagged as "failed" in the Variation pipeline.
The list of reasons for a variation being flagged as failed is available in the Quality Control documentation.
Contains alleles that did not pass the Ensembl filters
| See also: |
List of species with populated data:
|
This table contains descriptions of reasons for a variation being flagged as failed.
Example:
See below the list of the descriptions available in the Ensembl variation databases:
SELECT * FROM failed_description;
| See also: |
List of species with populated data:
|
For various reasons it may be necessary to store information about a structural variation that has failed quality checks (mappings) in the Structural Variation pipeline. This table acts as a flag for such failures.
| See also: |
List of species with populated data:
|
For various reasons it may be necessary to store information about a variation that has failed quality checks in the Variation pipeline. This table acts as a flag for such failures.
| See also: |
List of species with populated data:
|
For various reasons it may be necessary to store information about a variation feature that has failed quality checks. This table acts as a flag for such failures.
| See also: |
List of species with populated data:
|
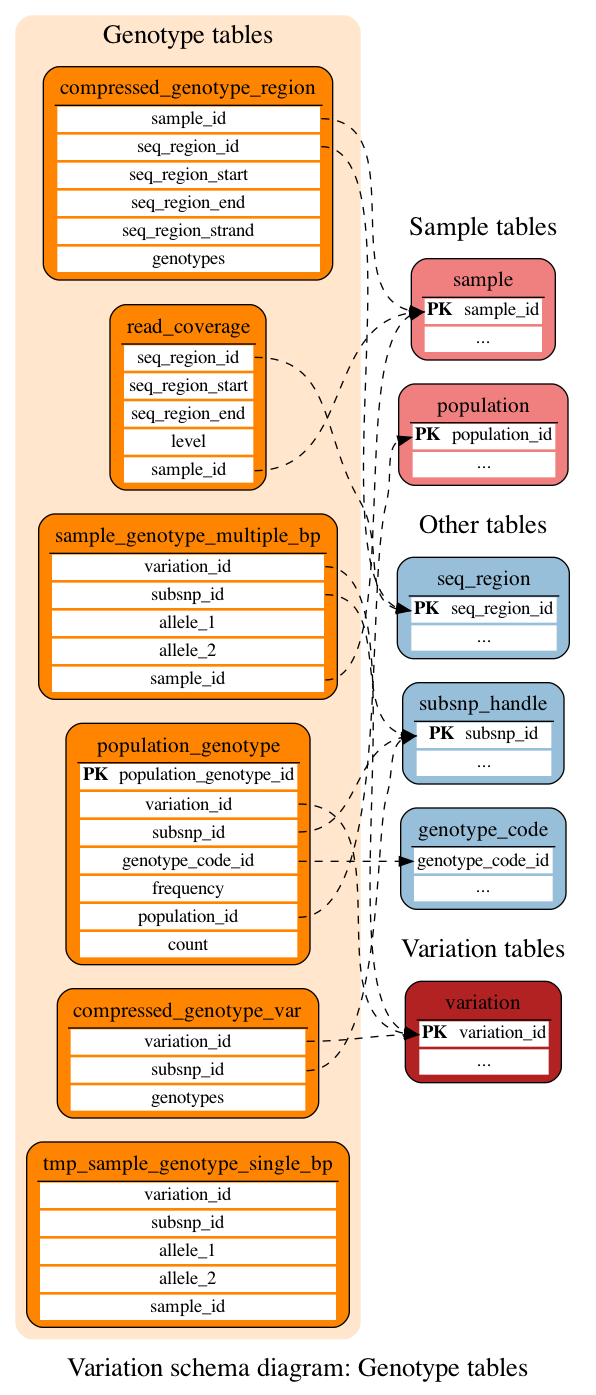
Genotype tables
These tables define the genotype data at the sample and population levels.
This table holds genotypes compressed using the pack() method in Perl. These genotypes are mapped to particular genomic locations rather than variation objects. The data have been compressed to reduce table size and increase the speed of the web code when retrieving strain slices and LD data. Only data from resequenced samples are used for LD calculations are included in this table
| See also: |
List of species with populated data:
|
This table holds genotypes compressed using the pack() method in Perl. These genotypes are mapped directly to variation objects. The data have been compressed to reduce table size. All genotypes in the database are included in this table (included duplicates of those genotypes contained in the compressed_genotype_region table). This table is optimised for retrieval from variation.
| See also: |
List of species with populated data:
|
This table stores genotypes and frequencies for variations in given populations.
| See also: |
List of species with populated data:
|
This table stores the read coverage of resequenced samples. Each row contains sample ID, chromosomal coordinates and a read coverage level.
| See also: |
List of species with populated data:
|
This table holds uncompressed genotypes for given variations.
| See also: |
List of species with populated data:
|
This table is only needed to create master schema when run healthcheck system. Needed for other species, but human, so keep it.
| See also: |
List of species with populated data:
|
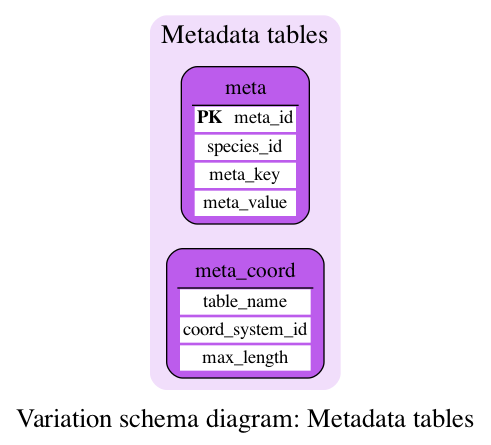
Metadata tables
These tables define some metadata information.
This table stores various metadata relating to the database, generally used by the Ensembl web code.
List of species with populated data:
|
This table gives the coordinate system used by various tables in the database.
List of species with populated data:
|
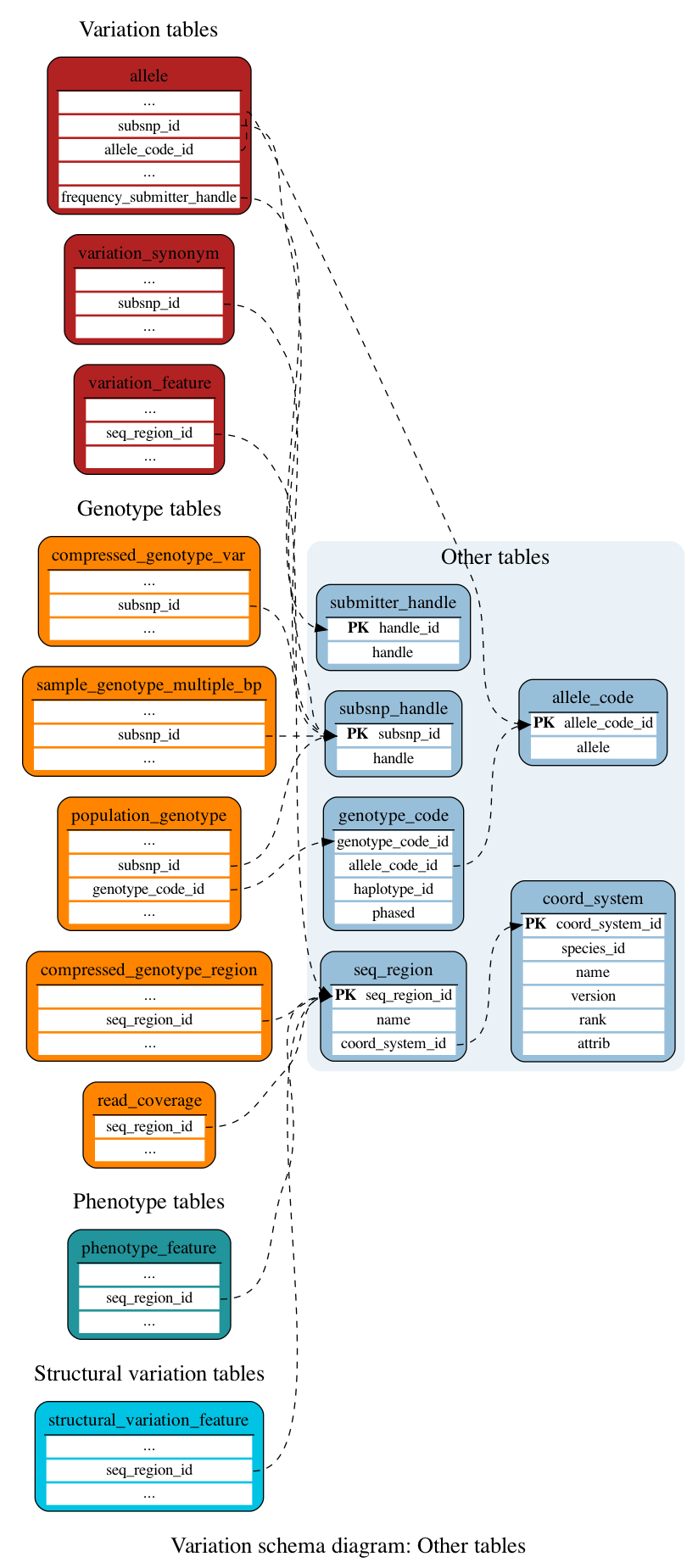
Other tables
These tables define the other data associated with a variation.
This table stores the relationship between the internal allele identifiers and the alleles themselves.
| See also: |
List of species with populated data:
|
Stores information about the available co-ordinate systems for the species identified through the species_id field. Note that for each species, there must be one co-ordinate system that has the attribute "top_level" and one that has the attribute "sequence_level".
| See also: |
List of species with populated data:
|
This table stores genotype codes as multiple rows of allele_code identifiers, linked by genotype_code_id and ordered by haplotype_id.
| See also: |
List of species with populated data:
|
This table stores the relationship between Ensembl's internal coordinate system identifiers and traditional chromosome names.
| See also: |
List of species with populated data:
|
This table holds a short string to distinguish data submitters
| See also: |
List of species with populated data:
|
This table contains the SubSNP(ss) ID and the name of the submitter handle of dbSNP.
| See also: |
List of species with populated data:
|
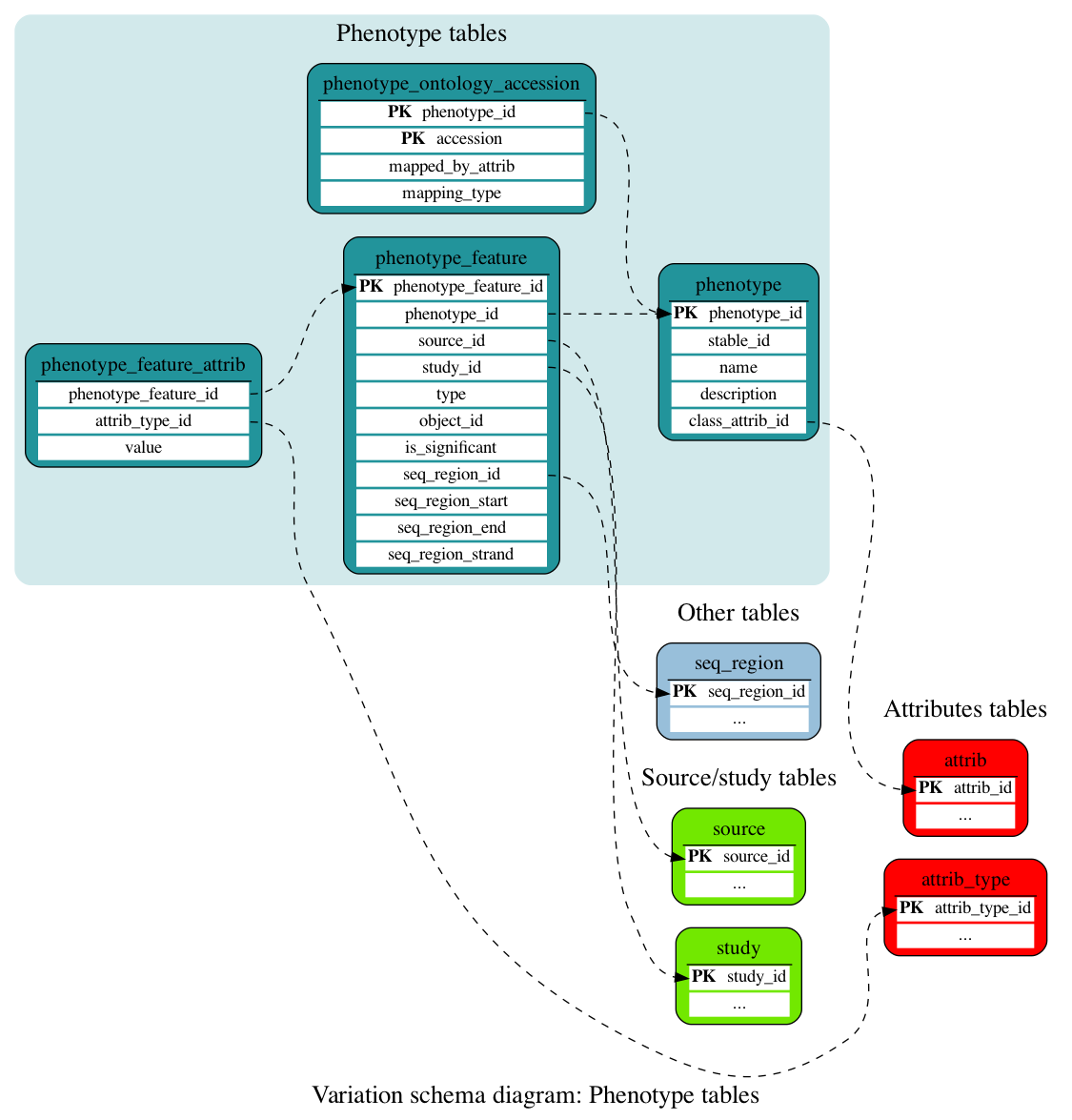
Phenotype tables
These tables store information linking entities (variants, genes, QTLs) with phenotypes and other annotations.
This table stores details of the phenotypes associated with phenotype_features.
| See also: |
List of species with populated data:
|
This table stores information linking entities (variants, genes, QTLs) and phenotypes.
| See also: |
List of species with populated data:
|
This table stores additional information on a given phenotype/object association. It is styled as an attrib table to allow for a variety of fields to be populated across different object types.
| See also: |
List of species with populated data:
|
This table stores accessions of phenotype ontology terms which have been linked to phenotype.descriptions
| See also: |
List of species with populated data:
|
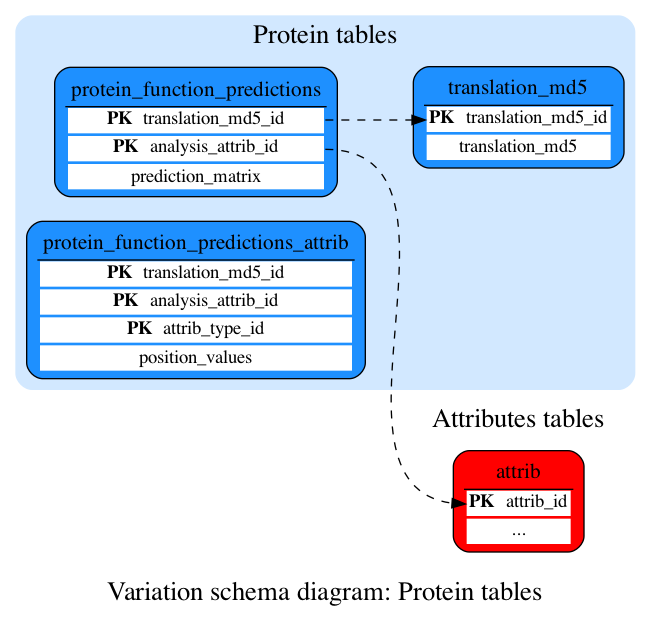
Protein tables
These tables define the protein prediction data.
Contains encoded protein function predictions for every protein-coding transcript in this species.
| See also: |
List of species with populated data:
|
Contains information on the data use in protein function predictions
| See also: |
List of species with populated data:
|
Maps a hex MD5 hash of a translation sequence to an ID used for the protein function predictions
| See also: |
List of species with populated data:
|
Sample tables
These tables define the sample, individual and population information.
Used to store groups of populations displayed separately on the Population Genetics page
| See also: |
List of species with populated data:
|
Stores information about an identifiable individual, including gender and the identifiers of the individual's parents (if known).
| See also: |
List of species with populated data:
|
Used to store alternative names for individuals when data comes from multiple sources.
| See also: |
List of species with populated data:
|
This table gives a detailed description for each of the possible individual types: fully_inbred, partly_inbred, outbred, mutant
Example:
See below the list of individual types:
SELECT * FROM individual_type;
| See also: |
List of species with populated data:
|
Stores information about a population. A population may be an ethnic group (e.g. Caucasian, Hispanic), assay group (e.g. 24 Europeans), phenotypic group (e.g. blue eyed, diabetes) etc. Populations may be composed of other populations by defining relationships in the population_structure table.
| See also: |
List of species with populated data:
|
This table stores hierarchical relationships between populations by relating them as populations and sub-populations.
| See also: |
List of species with populated data:
|
Used to store alternative names for populations when data comes from multiple sources.
| See also: |
List of species with populated data:
|
Stores information about a sample. A sample belongs to an individual. An individual can have multiple samples. A sample can belong only to one individual. A sample can be associated with a study.
| See also: |
List of species with populated data:
|
This table resolves the many-to-many relationship between the sample and population tables; i.e. samples may belong to more than one population. Hence it is composed of rows of sample and population identifiers.
| See also: |
List of species with populated data:
|
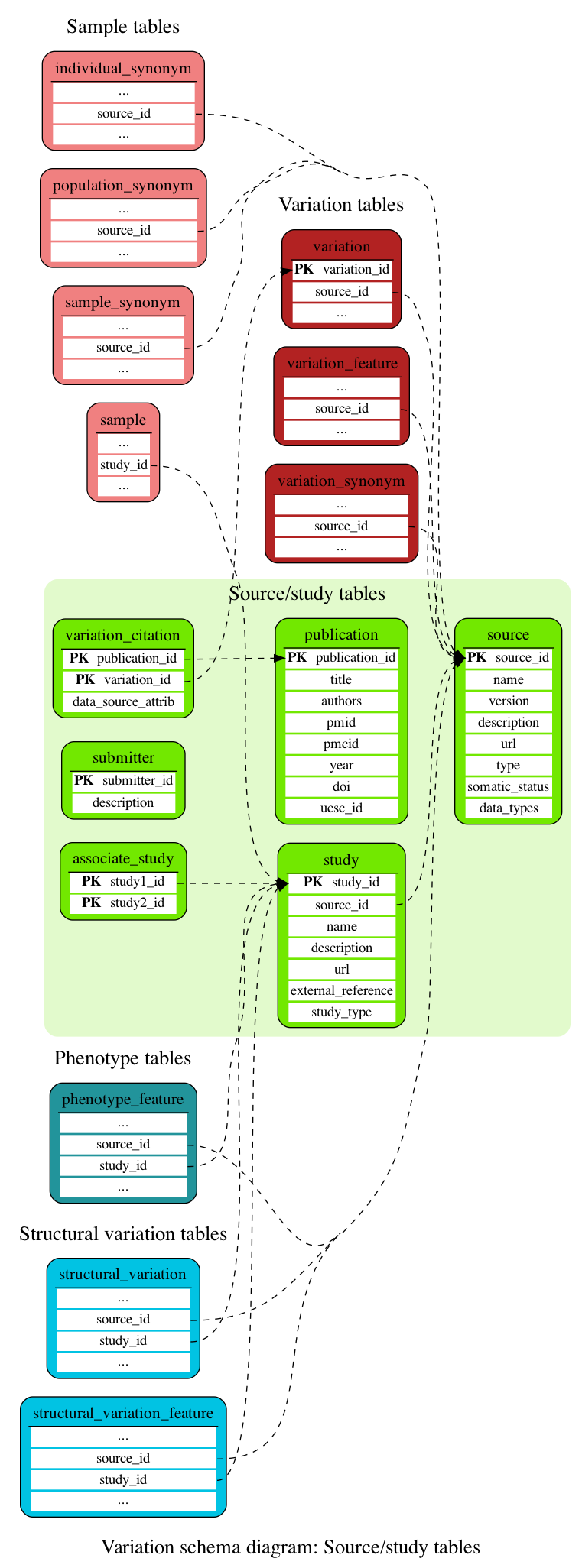
Source/study tables
These tables define the variation source and study information.
This table contains identifiers of associated studies (e.g. NHGRI and EGA studies with the same PubMed identifier).
| See also: |
List of species with populated data:
|
This table contains details of publications citing variations. This information comes from dbSNP, UCSC and Europe PMC.
| See also: |
List of species with populated data:
|
This table contains details of the source from which a variation is derived. Most commonly this is NCBI's dbSNP; other sources include SNPs called by Ensembl.
You can see the complete list, by species, here.
Example:
See below the command listing all the data sources in the human variation database:
SELECT * FROM source ORDER BY source_id;
| See also: |
List of species with populated data:
|
This table contains details of the studies. The studies information can come from internal studies (DGVa, EGA) or from external studies (UniProt, NHGRI, ...).
| See also: |
List of species with populated data:
|
This table contains descriptions of group submitting data to public repositories such as ClinVar
| See also: |
List of species with populated data:
|
This table links a variation to a publication
| See also: |
List of species with populated data:
|
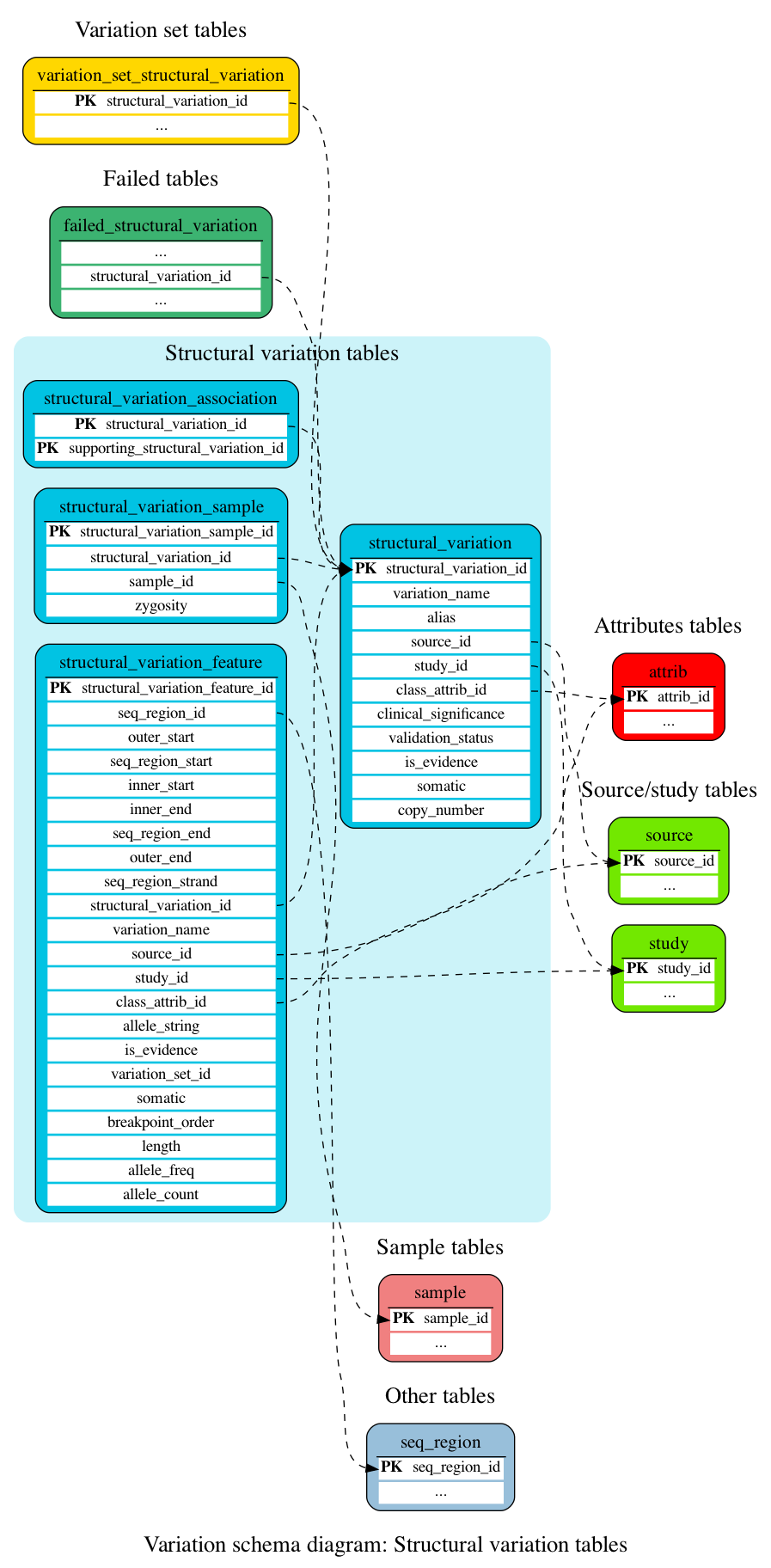
Structural variation tables
These tables define the structural variation data.
This table stores information about structural variation.
| See also: |
List of species with populated data:
|
This table stores the associations between structural variations and their supporting evidences.
| See also: |
List of species with populated data:
|
This table stores information about structural variation features (i.e. mappings of structural variations to genomic locations).
| See also: |
List of species with populated data:
|
This table stores sample and strain information for structural variants and their supporting evidences.
| See also: |
List of species with populated data:
|
Variation effect tables
These tables define the variation effect prediction data in different Ensembl features.
This table relates a single allele of a variation_feature to a motif feature (see Regulation documentation). It contains the consequence of the allele.
| See also: |
List of species with populated data:
|
This table relates a single allele of a variation_feature to a regulatory feature (see Regulation documentation). It contains the consequence of the allele.
| See also: |
List of species with populated data:
|
This table relates a single allele of a variation_feature to a transcript (see Core documentation). It contains the consequence of the allele e.g. intron_variant, non_synonymous_codon, stop_lost etc, along with the change in amino acid in the resulting protein if applicable.
| See also: |
List of species with populated data:
|
This table is used in web index creation. It links a variation_id to the names of the genes the variation is within
| See also: |
List of species with populated data:
|
This table is used in web index creation. It links a variation_id to all possible transcript and protein level change descriptions in HGVS annotation.
| See also: |
List of species with populated data:
|
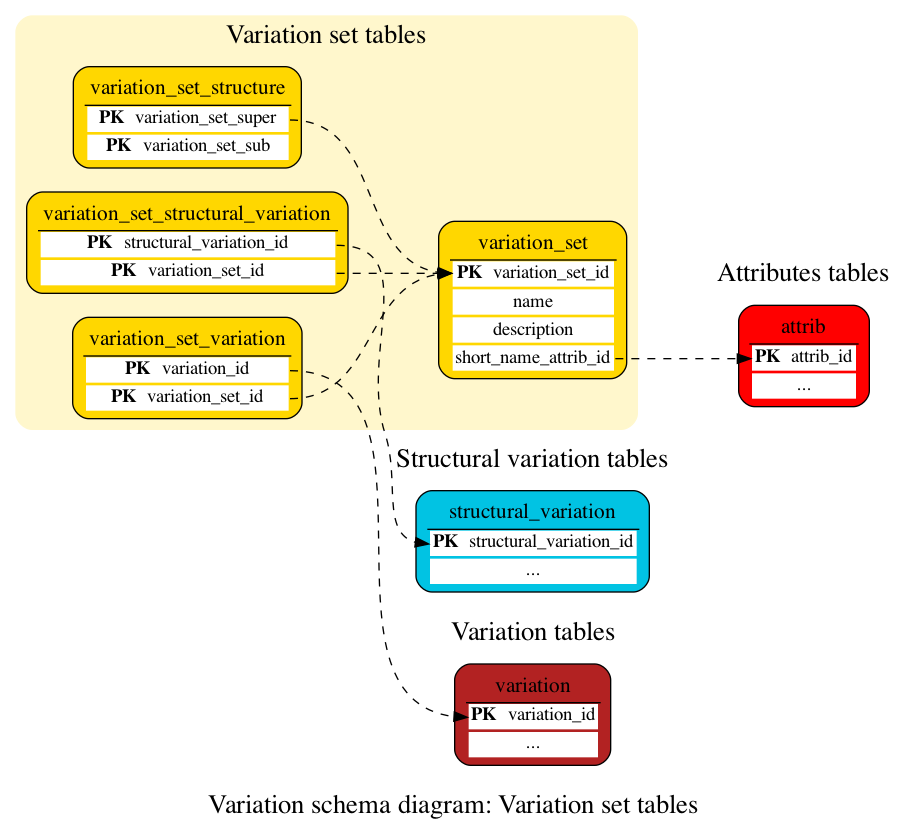
Variation set tables
These tables define the variation and structural variation set data. The complete list of variation sets with their descriptions is available here.
This table contains the name of sets and subsets of variations stored in the database. It usually represents the name of the project or subproject where a group of variations has been identified.
Example:
See below the command to display the list of variation set entries, e.g. for human:
SELECT * FROM variation_set;
| See also: |
List of species with populated data:
|
A table for mapping structural variations to variation_sets.
| See also: |
List of species with populated data:
|
This table stores hierarchical relationships between variation sets by relating them as variation sets and variation subsets.
| See also: |
List of species with populated data:
|
A table for mapping variations to variation_sets.
| See also: |
List of species with populated data:
|
Variation tables
These tables define the central variation data.
This table stores information about each of a variation's alleles, along with population frequencies.
| See also: |
List of species with populated data:
|
This table allows for the allele of a variant to have multiple IDs.
| See also: |
List of species with populated data:
|
This is the schema's generic representation of a variation, defined as a genetic feature that varies between individuals of the same species. The most common type is the single nucleotide variation (SNP) though the schema also accommodates copy number variations (CNVs) and structural variations (SVs).
In Ensembl, a variation is defined by its flanking sequence rather than its mapped location on a chromosome; a variation may in fact have multiple mappings across a genome, although this fails our Quality Control.
This table stores a variation's name (commonly an ID of the form e.g. rs123456, assigned by dbSNP).
| See also: |
List of species with populated data:
|
This table stores miscellaneous attributes associated with a variation entry.
| See also: |
List of species with populated data:
|
This table represents mappings of variations to genomic locations. It stores an allele string representing the different possible alleles that are found at that locus e.g. "A/T" for a SNP, as well as a "worst case" consequence of the mutation. It also acts as part of the relationship between variations and transcripts.
| See also: |
List of species with populated data:
|
This table allows for a variation to have multiple IDs, generally given by multiple sources.
| See also: |
List of species with populated data:
|


 Show columns
Show columns