Ensembl Compara Schema Documentation
Introduction
This document describes the tables that make up the Ensembl Compara schema. Tables are listed grouped in different categories, and the purpose of each table is explained. Several examples are also given. They are intended to allow people to familiarise themselves with the schema. The overall diagram can be found here.
List of the tables:
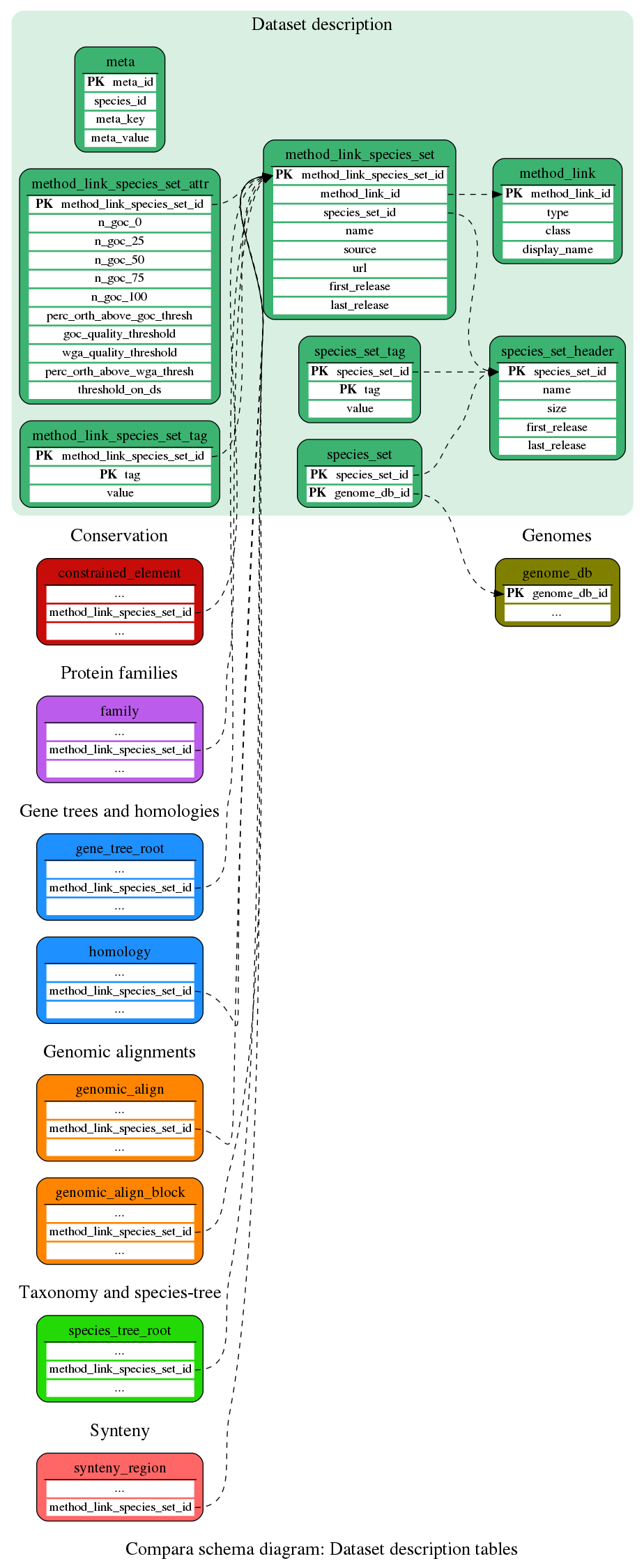
Dataset description
These are general tables used in the Compara schema
This table stores meta information about the compara database
Example:
This query defines which API version must be used to access this database.
SELECT * FROM meta WHERE meta_key = "schema_version";
Header for the species_set table which groups or sets of species which are used in the method_link_species_set table.
Example:
This query shows the first 10 species_sets having human
SELECT ssh.species_set_id, ssh.name, ssh.size, gdb.genome_db_id, gdb.name, gdb.assembly FROM species_set_header ssh JOIN species_set ss USING(species_set_id) JOIN genome_db gdb USING(genome_db_id) WHERE ssh.name="primates";
| See also: |
Describes the content of each species-set (species_set_header) as a set of genome_db objects
Example:
This query shows the first 10 species_sets having human
SELECT species_set_id, GROUP_CONCAT(name) AS species FROM species_set JOIN genome_db USING(genome_db_id) GROUP BY species_set_id HAVING species LIKE '%homo_sapiens%' ORDER BY species_set_id LIMIT 10;
| See also: |
This table contains descriptive tags for the species_set_ids in the species_set table. It is used to store options on clades and group of species. It has been initially developed for the gene tree view.
| See also: |
This table specifies which kind of link can exist between entities in compara (dna/dna alignment, synteny regions, homologous gene pairs, etc...)
NOTE: We use method_link_ids between 1 and 100 for DNA-DNA alignments, between 101 and 200 for genomic syntenies, between 201 and 300 for protein homologies, between 301 and 400 for protein families and between 401 and 500 for protein and ncRNA trees. Each category corresponds to data stored in different tables.
Example:
These are our current entries:
SELECT * FROM method_link;
| See also: |
This table contains information about the comparisons stored in the database. A given method_link_species_set_id exist for each comparison made and relates a method_link_id in method_link with a set of species (species_set_id) in the species_set table.
Example:
This query shows all the EPO alignments in this database:
SELECT * FROM method_link_species_set WHERE method_link_id = 13;
| See also: |
Contains serveral tag/value data associated with method_link_species_set entries
| See also: |
This table contains the distribution of the gene order conservation scores
| See also: |
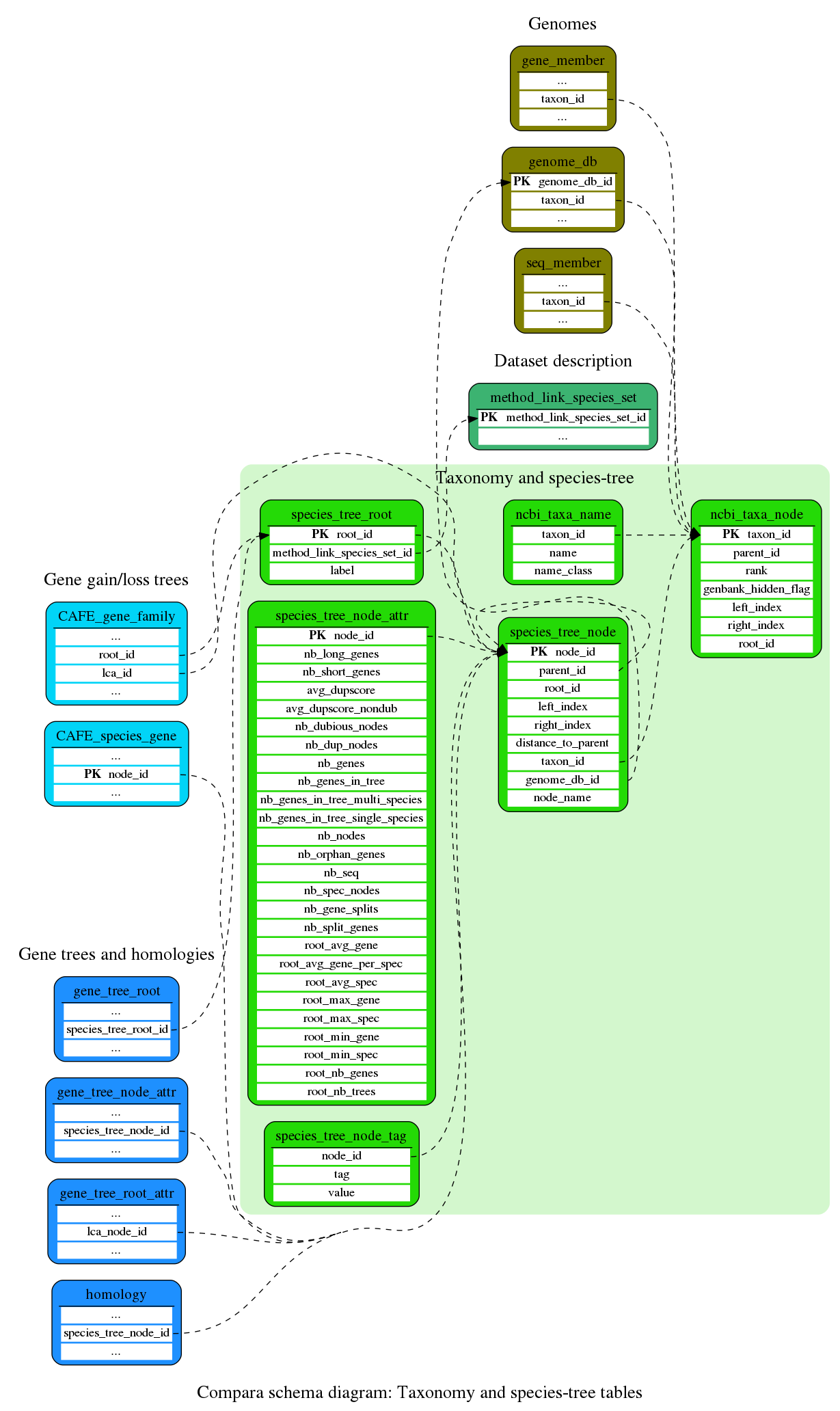
Taxonomy and species-tree
Species-tree used in the Compara analyses (incl. new annotations generated in-house), and the NCBI taxonomy (which often used as a template for species-trees)
This table contains all taxa used in this database, which mirror the data and tree structure from NCBI Taxonomy database (for more details see ensembl-compara/script/taxonomy/README-taxonomy which explain our import process)
Example:
This examples shows how to get the lineage for Homo sapiens:
SELECT n2.taxon_id, n2.parent_id, na.name, n2.rank, n2.left_index, n2.right_index FROM ncbi_taxa_node n1 JOIN (ncbi_taxa_node n2 LEFT JOIN ncbi_taxa_name na ON n2.taxon_id = na.taxon_id AND na.name_class = "scientific name") ON n2.left_index <= n1.left_index AND n2.right_index >= n1.right_index WHERE n1.taxon_id = 9606 ORDER BY left_index;
| See also: |
This table contains different names, aliases and meta data for the taxa used in Ensembl.
Example:
Here is an example on how to get the taxonomic ID for a species:
SELECT * FROM ncbi_taxa_name WHERE name_class = "scientific name" AND name = "Homo sapiens";
| See also: |
This table contains the nodes of the species tree used in the gene gain/loss analysis
| See also: |
This table stores species trees used in compara. Each tree is made of species_tree_node's
Example:
Retrieve all the species trees stored in the database
SELECT * FROM species_tree_root
| See also: |
This table contains tag/value data for species_tree_nodes
| See also: |
This table contains tag/value data for species_tree_nodes
| See also: |
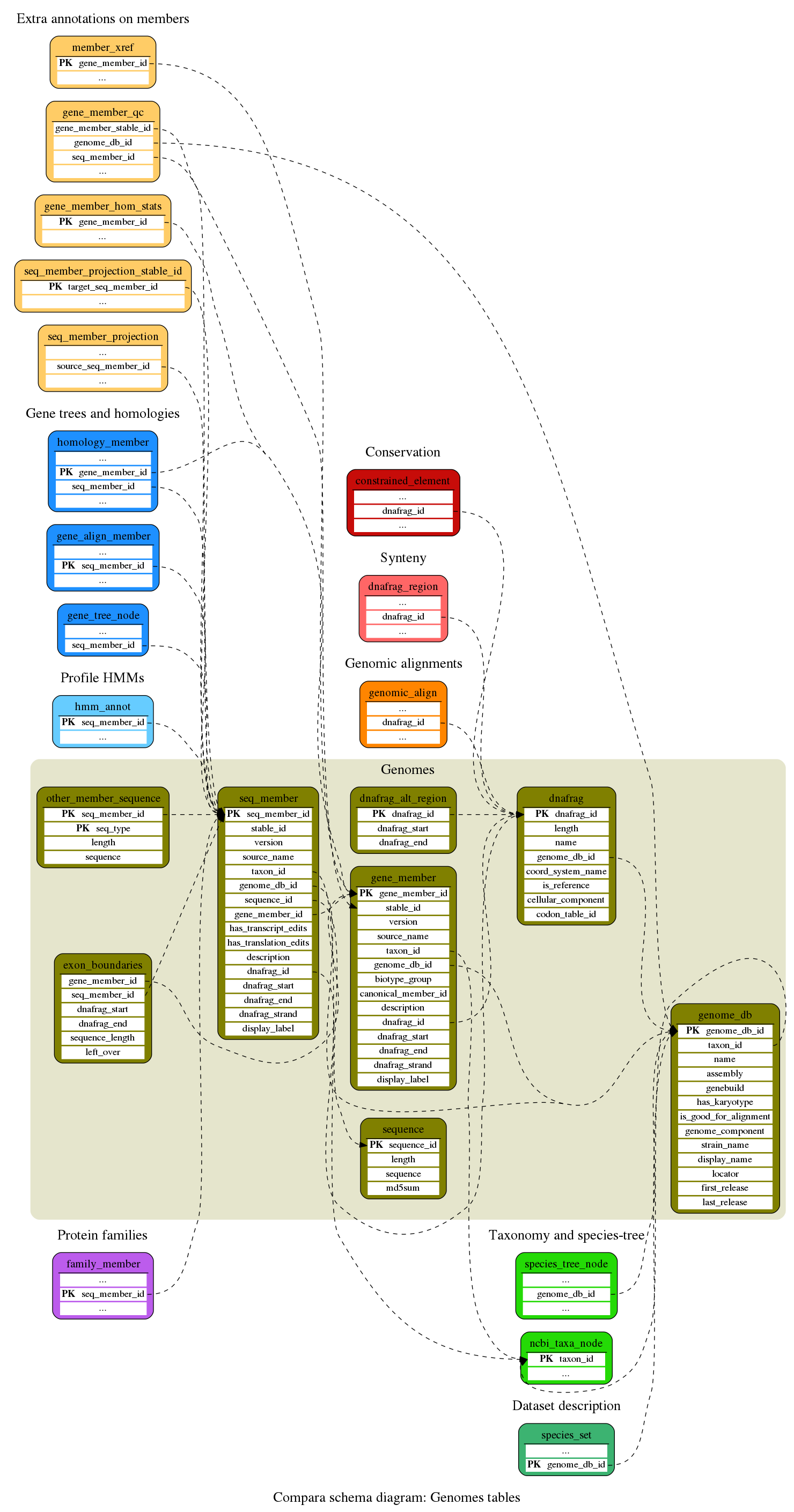
Genomes
Description of the genomes (assembly, sequences, genes, etc)
This table contains information about the version of the genome assemblies used in this database
Example:
This query shows the entries for human and chicken
SELECT * FROM genome_db WHERE name IN ("Homo_sapiens", "Gallus_gallus");
| See also: |
This table defines the genomic sequences used in the comparative genomics analyisis. It is used by the genomic_align_block table to define aligned sequences. It is also used by the dnafrag_region table to define syntenic regions.
NOTE: Index <name> has genome_db_id in the first place because unless fetching all dnafrags or fetching by dnafrag_id, genome_db_id appears always in the WHERE clause. Unique key <name> is used to ensure that Bio::EnsEMBL::Compara::DBSQL::DnaFragAdaptor->fetch_by_GenomeDB_and_name will always fetch a single row. This can be used in the EnsEMBL Compara DB because we store top-level dnafrags only.
Example:
This query shows the chromosome 14 of the Human genome (genome_db.genome_db_id = 150 refers to Human genome in this example) which is 107349540 nucleotides long.
SELECT dnafrag.* FROM dnafrag LEFT JOIN genome_db USING (genome_db_id) WHERE dnafrag.name = "14" AND genome_db.name = "homo_sapiens";
| See also: |
This table lists the position of the alternative sequences on non-reference dnafrags (haplotypes and assembly patches). On these dnafrags, only the region between these two coordinates differ from the base (reference) dnafrag.
Example:
This query lists some haplotypes of the human chromosome 20, incl. the length of the complete patched chromosome and the coordinates of the region that differs.
SELECT dnafrag_id, name, length, dnafrag_start, dnafrag_end FROM dnafrag_alt_regiON JOIN dnafrag USING (dnafrag_id) WHERE name LIKE "CHR\_HSCHR20\_%";
| See also: |
This table contains the sequences of the seq_member entries
This table links sequences to the EnsEMBL core DB or to external DBs.
Example:
The following query refers to the human (ncbi_taxa_node.taxon_id = 9606 or genome_db_id = 150) gene ENSG00000176105
SELECT * FROM gene_member WHERE stable_id = "ENSG00000176105";
| See also: |
This table links sequences to the EnsEMBL core DB or to external DBs.
Example:
The following query refers to the human (ncbi_taxa_node.taxon_id = 9606 or genome_db_id = 150) peptide ENSP00000324740
SELECT * FROM seq_member WHERE stable_id = "ENSP00000324740";
| See also: |
This table stores the exon coordinates of a seq_member. Coordinates are assumed to be on the dnafrag of the seq_member
| See also: |
This table includes alternative sequences for Member, like sequences with flanking regions
| See also: |
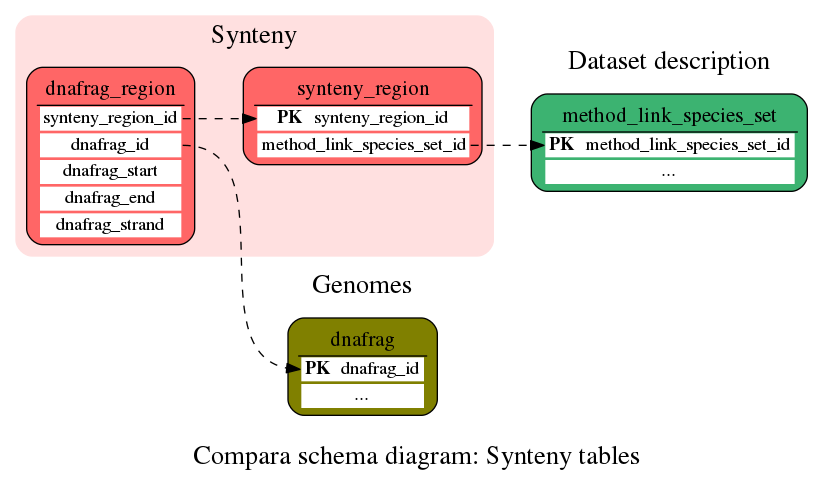
Synteny
These tables store information about genomic alignments in the Compara schema
Contains all the syntenic relationships found and the relative orientation of both syntenic regions.
Example:
This query shows the 4 first syntenic regions between the Human and Opossum genomes by linking with the method_link_species_set table
SELECT synteny_region.* FROM synteny_regiON JOIN method_link_species_set USING (method_link_species_set_id) JOIN species_set_header USING (species_set_id) WHERE species_set_header.name = "Hsap-Mdom" LIMIT 4;
| See also: |
This table contains the genomic regions corresponding to every synteny relationship found. There are two genomic regions for every synteny relationship.
Example 1:
Return two top dnafrag regions
SELECT * FROM dnafrag_regiON ORDER BY synteny_region_id LIMIT 2;
Example 2:
Same two dnafrag regions, but annotated
SELECT genome_db.name AS species_name, dnafrag.name, dnafrag_start, dnafrag_end, dnafrag_strAND FROM dnafrag_regiON JOIN dnafrag USING (dnafrag_id) JOIN genome_db USING (genome_db_id) ORDER BY synteny_region_id LIMIT 2;
| See also: |
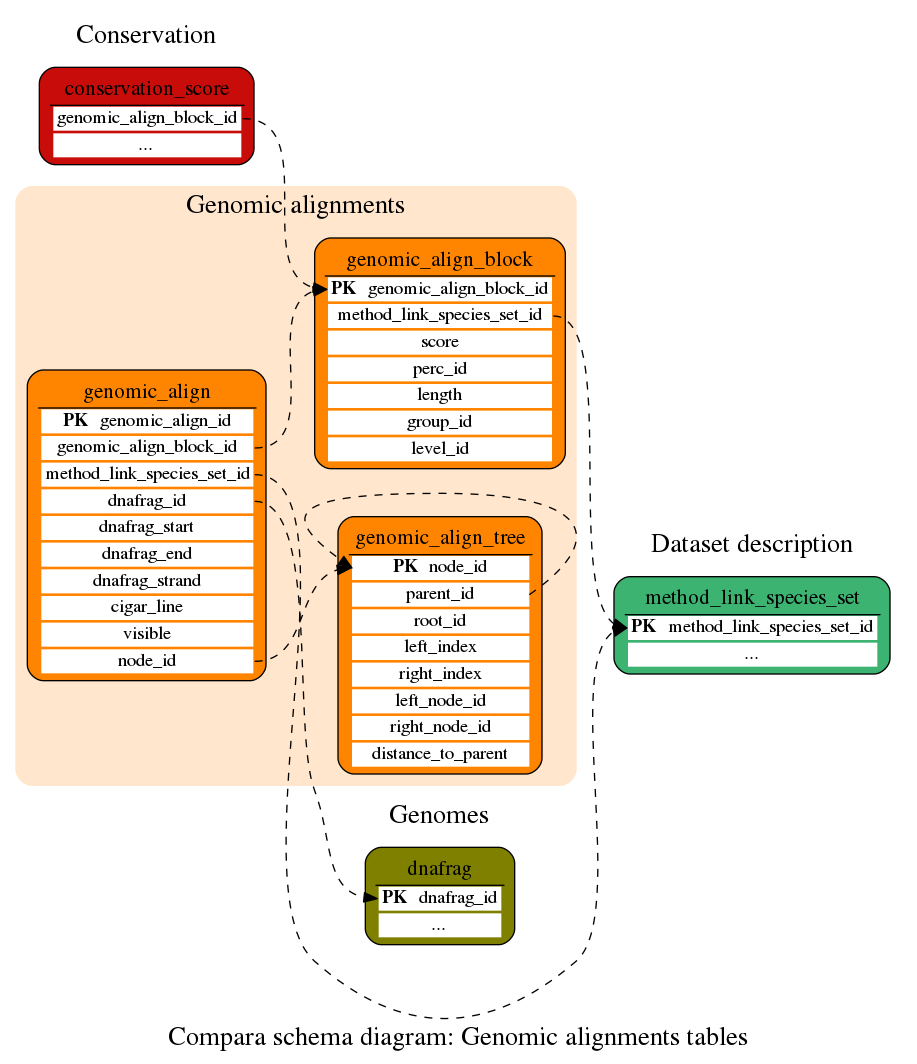
Genomic alignments
Whole-genome alignments (as blocks and trees)
This table is the key table for the genomic alignments. The software used to align the genomic blocks is refered as an external key to the method_link table. Nevertheless, actual aligned sequences are defined in the genomic_align table.
Tree alignments (EPO alignments) are best accessed through the genomic_align_tree table although the alignments are also indexed in this table. This allows the user to also access the tree alignments as normal multiple alignments.
NOTE: All queries in the API uses the primary key as rows are always fetched using the genomic_align_block_id. The key 'method_link_species_set_id' is used by MART when fetching all the genomic_align_blocks corresponding to a given method_link_species_set_id
Example:
The following query refers to the LastZ alignment between medaka and zebrafish:
SELECT genomic_align_block.* FROM genomic_align_block JOIN method_link_species_set USING (method_link_species_set_id) JOIN species_set_header USING (species_set_id) WHERE method_link_id = 16 AND species_set_header.name = "Drer-Olat" ORDER BY genomic_align_block_id LIMIT 4;
| See also: |
This table is used to index tree alignments, e.g. EPO alignments. These alignments include inferred ancestral sequences. The tree required to index these sequences is stored in this table. This table stores the structure of the tree. Each node links to an entry in the genomic_align_group table, which links to one or several entries in the genomic_align table.
NOTE: Left_index and right_index are used to speed up fetching trees from the database. Any given node has its left_index larger than the left_index of its parent node and its right index smaller than the right_index of its parent node. In other words, all descendent nodes of a given node can be obtained by fetching all the node with a left_index (or right_index or both) between the left_index and the right_index of that node.
Example 1:
The following query corresponds to the root of a tree, because parent_id = 0 and root_id = node_id
SELECT * FROM genomic_align_tree WHERE node_id = root_id LIMIT 1;
Example 2:
Grab the first two trees that have exactly 5 nodes
SELECT * FROM genomic_align_tree WHERE root_id IN (SELECT root_id FROM genomic_align_tree GROUP BY root_id HAVING COUNT(*) = 5) LIMIT 10;
| See also: |
This table contains the coordinates and all the information needed to rebuild genomic alignments. Every entry corresponds to one of the aligned sequences. It also contains an external key to the method_link_species_set which refers to the software and set of species used for getting the corresponding alignment. The aligned sequence is defined by an external reference to the dnafrag table, the starting and ending position within this dnafrag, the strand and a cigar_line.
The original aligned sequence is not stored but it can be retrieved using the cigar_line field and the original sequence. The cigar line defines the sequence of matches/mismatches and deletions (or gaps). For example, this cigar line 2MD3M2D2M will mean that the alignment contains 2 matches/mismatches, 1 deletion (number 1 is omitted in order to save some space), 3 matches/mismatches, 2 deletions and 2 matches/mismatches. If the original sequence is AACGCTT, the aligned sequence will be:
| M | M | D | M | M | M | D | D | M | M |
|---|---|---|---|---|---|---|---|---|---|
| A | A | - | C | G | C | - | - | T | T |
Example 1:
The following query corresponds to the 4x2 sequences included in the alignment described above (see genomic_align_block table description).
SELECT genomic_align.* FROM genomic_align_block JOIN method_link_species_set USING (method_link_species_set_id) JOIN species_set_header USING (species_set_id) JOIN genomic_align USING (genomic_align_block_id) WHERE method_link_id = 16 AND species_set_header.name = "Drer-Olat" ORDER BY genomic_align_block_id LIMIT 8;
Example 2:
Here is a better way to get this by joining the dnafrag and genome_db tables:
SELECT genome_db.name, dnafrag.name, dnafrag_start, dnafrag_end, dnafrag_strAND str, cigar_line FROM genomic_align_block JOIN method_link_species_set USING (method_link_species_set_id) JOIN species_set_header USING (species_set_id) JOIN genomic_align USING (genomic_align_block_id) JOIN dnafrag USING (dnafrag_id) JOIN genome_db USING (genome_db_id) WHERE method_link_id = 16 AND species_set_header.name = "Drer-Olat" ORDER BY genomic_align_block_id LIMIT 8;
| See also: |
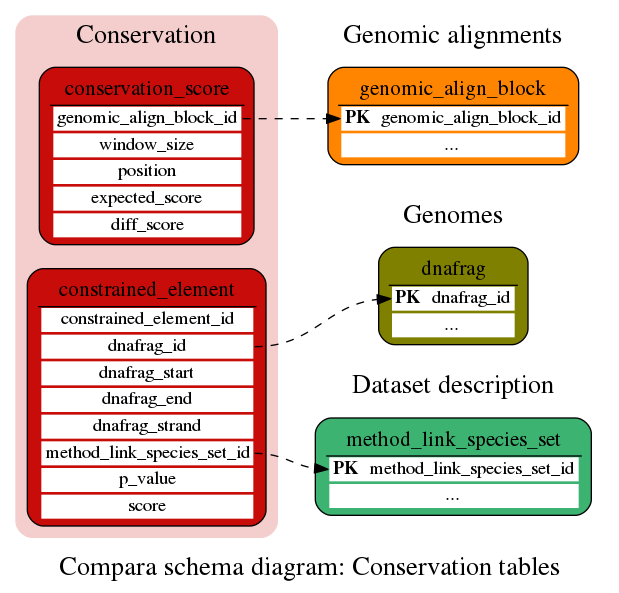
Conservation
Evolutionary conservation (scores and regions)
This table contains conservation scores calculated from the whole-genome multiple alignments stored in the genomic_align_block table. Several scores are stored per row. expected_score and diff_score are binary columns and you need to use the Perl API to access these data.
| See also: |
This table contains constrained elements calculated from the whole-genome multiple alignments stored in the genomic_align_block table
Example 1:
Example entry for a constrained_element:
SELECT * FROM constrained_element ORDER BY constrained_element_id LIMIT 1;
Example 2:
There are 2 other elements in the same constrained_element:
SELECT * FROM constrained_element JOIN (SELECT MIN(constrained_element_id) AS constrained_element_id FROM constrained_element) t USING (constrained_element_id);
| See also: |
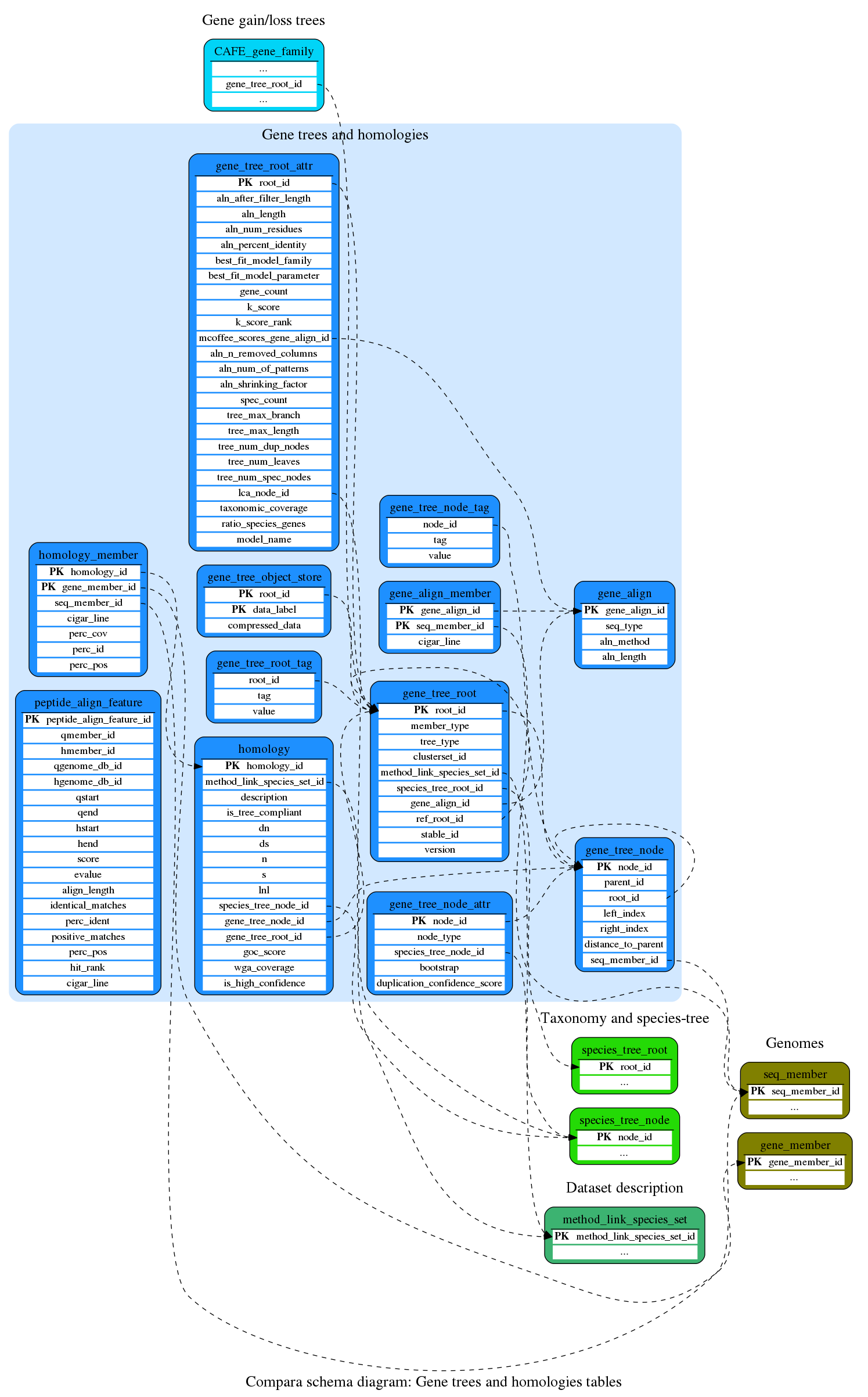
Gene trees and homologies
These tables store information about gene alignments, trees and homologies
: This table stores the raw local alignment results of peptide to peptide alignments returned by a BLAST run. The hits are actually stored in species-specific tables rather than in a single table. For example, human has the genome_db_id 150, and all the hits that have a human gene as a query are stored in peptide_align_feature
Example 1:
Example of peptide_align_feature entry: sql SELECT * FROM peptide_align_feature WHERE hgenome_db_id = 111 LIMIT 1;
Example 2:
The following query corresponds to a particular hit found between a Homo sapiens protein and a Anolis carolinensis protein: sql SELECT g1.name as qgenome, m1.stable_id as qstable_id, g2.name as hgenome, m2.stable_id as hstable_id, score, evalue FROM peptide_align_feature JOIN seq_member m1 ON (qmember_id = m1.seq_member_id) JOIN seq_member m2 ON (hmember_id = m2.seq_member_id) JOIN genome_db g1 ON (qgenome_db_id = g1.genome_db_id) JOIN genome_db g2 ON (hgenome_db_id = g2.genome_db_id) WHERE hgenome_db_id = 111 LIMIT 1;
| See also: |
This table stores information about alignments for members
| See also: |
This table allows certain nodes (leaves) to have aligned protein member_scores attached to them
| See also: |
This table holds the gene tree data structure, such as root, relation between parent and child, leaves, etc... In our data structure, all the trees of a given clusterset are arbitrarily connected to the same root. This eases to store and query in the same database the data from independant tree building analysis. Hence the "biological roots" of the trees are the children nodes of the main clusterset root. See the examples below.
Example:
The following query returns the root nodes of the independant protein trees stored in the database
SELECT gtn.node_id FROM gene_tree_node gtn LEFT JOIN gene_tree_root gtr ON (gtn.parent_id = gtr.root_id) WHERE gtr.tree_type = 'clusterset' AND gtr.member_type = 'protein' LIMIT 10;
| See also: |
Header table for gene_trees. The database is able to contain several sets of trees computed on the same genes. We call these analysis "clustersets" and they can be distinguished with the clusterset_id field. Traditionally, the compara databases have contained only one clusterset (clusterset_id=1), but currently (starting on release 66) we have at least 2 (one for protein trees and one for ncRNA trees). See the examples below.
Example 1:
The following query retrieves all the node_id of the current clustersets
SELECT * FROM gene_tree_root WHERE tree_type = 'clusterset';
Example 2:
To get the number of trees of each type
SELECT member_type, tree_type, COUNT(*) FROM gene_tree_root GROUP BY member_type, tree_type;
| See also: |
This table contains several tag/value data attached to node_ids
| See also: |
This table contains several tag/value data for gene_tree_roots
| See also: |
This table contains several gene tree attributes data attached to root_ids
| See also: |
This table contains several gene tree attributes data attached to node_ids
| See also: |
This table contains arbitrary data related to gene-trees. Commonly used for precomputed tracks / layers
| See also: |
This table contains all the genomic homologies. There are two homology_member entries for each homology entry for now, but both the schema and the API can handle more than just pairwise relationships.
dN, dS, N, S and lnL are statistical values given by the codeml program of the Phylogenetic Analysis by Maximum Likelihood (PAML) package.
Example 1:
The following query defines a pair of paralogous xenopous genes. See homology_member for more details
SELECT homology.* FROM homology JOIN method_link_species_set USING (method_link_species_set_id) WHERE name="Xtro paralogues" LIMIT 1;
Example 2:
See species_names that participate in this particular homology entry
SELECT homology_id, description, GROUP_CONCAT(genome_db.name) AS species FROM homology JOIN method_link_species_set USING (method_link_species_set_id) JOIN species_set USING (species_set_id) JOIN genome_db USING(genome_db_id) WHERE method_link_id=201 AND homology_id<200000000 GROUP BY homology_id LIMIT 4;
| See also: |
This table contains the sequences corresponding to every genomic homology relationship found. There are two homology_member entries for each pairwise homology entry. As written in the homology table section, both schema and API can deal with more than pairwise relationships.
The original alignment is not stored but it can be retrieved using the cigar_line field and the original sequences. The cigar line defines the sequence of matches or mismatches and deletions in the alignment.
- First peptide sequence: SERCQVVVISIGPISVLSMILDFY
- Second peptide sequence: SDRCQVLVISILSMIGLDFY
- First corresponding cigar line: 20MD4M
- Second corresponding cigar line: 11M5D9M
| First peptide cigar line | M | M | M | M | M | M | M | M | M | M | M | M | M | M | M | M | M | M | M | M | D | M | M | M | M |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| First aligned peptide | S | E | R | C | Q | V | V | V | I | S | I | G | P | I | S | V | L | S | M | I | - | L | D | F | Y |
| Second aligned peptide | S | D | R | C | Q | V | L | V | I | S | I | - | - | - | - | - | L | S | M | I | G | L | D | F | Y |
| Second peptide cigar line | M | M | M | M | M | M | M | M | M | M | M | D | D | D | D | D | M | M | M | M | M | M | M | M | M |
Example:
The following query refers to the two homologue sequences from the first xenopus' paralogy object. Gene and peptide sequence of the second homologue can retrieved in the same way.
SELECT homology_member.* FROM homology_member JOIN homology USING (homology_id) JOIN method_link_species_set USING (method_link_species_set_id) WHERE name="Xtro paralogues" LIMIT 2;
| See also: |
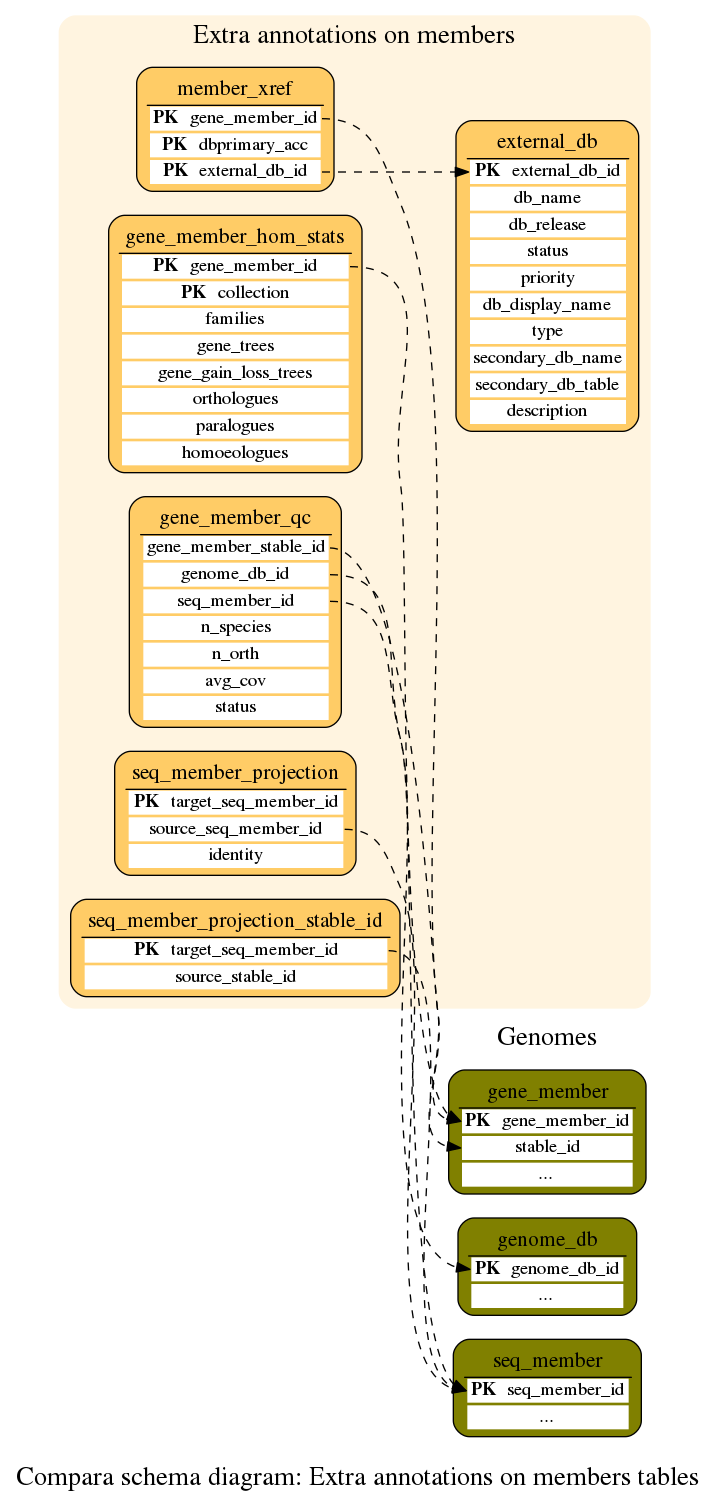
Extra annotations on members
Various member (gene and proteins) related information stored in the database, either loaded from Core databases or aggregated from Compara analyses
This table contains for each gene_member some statistics about the homology pipelines we've run
| See also: |
This table stores data about projected transcripts (in the gene-annotation process), which is used to help the clustering. This table links to the source stable_id and is used until the source members are loaded
Example:
The following query shows the projections of the mouse gene Pdk3 to all the other species
SELECT ss.stable_id, gs.name, source_stable_id FROM seq_member ss JOIN genome_db gs USING (genome_db_id) JOIN seq_member_projection_stable_id ON seq_member_id = target_seq_member_id WHERE source_stable_id = "ENSMUST00000045748"
| See also: |
This table stores data about projected transcripts (in the gene-annotation process), which is used to help the clustering. This table can only be used when both genomes have been loaded. Thus we first populate seq_member_projection_stable_id and then copy the data whilst transforming the stable_id into a seq_member_id
Example:
The following query shows the projections of the gene AGO2 with the Anole lizard and the zebrafinch sql SELECT ss.stable_id, gs.name, st.stable_id, gt.name, identity FROM seq_member ss JOIN genome_db gs USING (genome_db_id) JOIN seq_member_projection ON ss.seq_member_id = source_seq_member_id JOIN (seq_member st JOIN genome_db gt USING (genome_db_id)) ON st.seq_member_id=target_seq_member_id WHERE ss.stable_id IN ("ENSACAP00000000183", "ENSTGUP00000014905");
| See also: |
This table stores data about the external databases in which the objects described in the member_xref table are stored.
| See also: |
This table stores cross-references for gene members derived from the core databases. It is used by Bio::EnsEMBL::Compara::DBSQL::XrefMemberAdaptor and provides the data used in highlighting gene trees by GO and InterPro annotation"
| See also: |
This table contains gene quality information from the geneset_QC pipeline
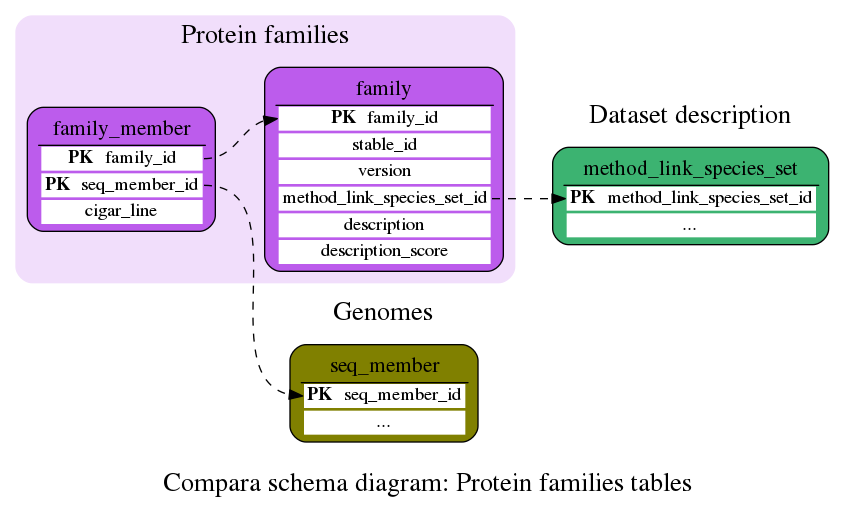
Protein families
Protein families (sets of homologous protein sequences)
This table contains all the group homologies found. There are several family_member entries for each family entry.
| See also: |
This table contains the proteins corresponding to protein family relationship found. There are several family_member entries for each family entry
| See also: |
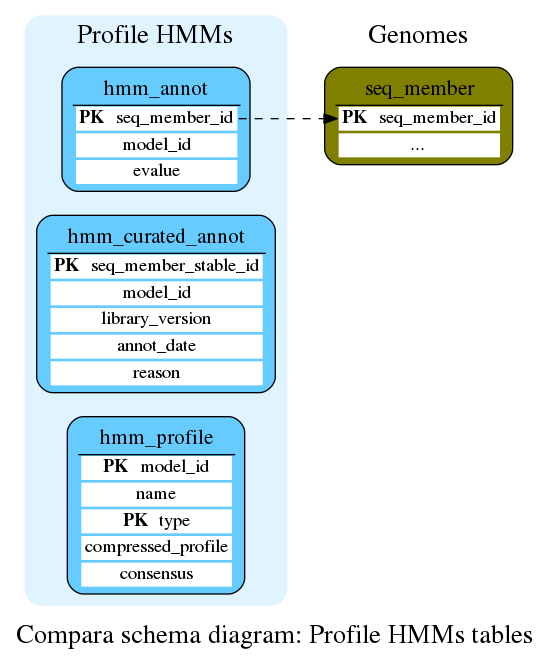
Profile HMMs
The profile HMMs and their hits
This table stores different HMM-based profiles used and produced by gene trees
This table stores the HMM annotation of the seq_members
| See also: |
This table stores the curated / forced HMM annotation of the seq_members
| See also: |
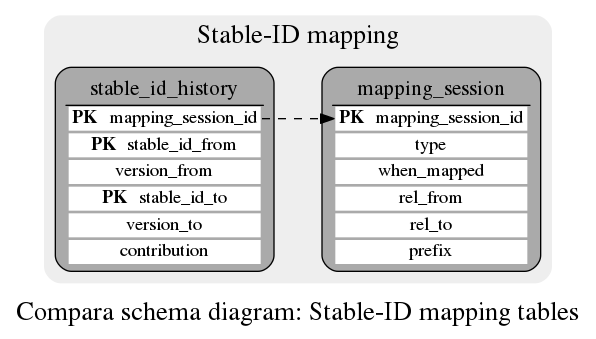
Stable-ID mapping
History of the gene-tree and family IDs across different versions of Ensembl
This table contains one entry per stable_id mapping session (either for Families or for Protein Trees), which contains the type, the date of the mapping, and which releases were linked together. A single mapping_session is the event when mapping between two given releases for a particular class type ('family' or 'tree') is loaded. The whole event is thought to happen momentarily at 'when_mapped' (used for sorting in historical order).
This table keeps the history of stable_id changes from one release to another. The primary key 'object' describes a set of members migrating from stable_id_from to stable_id_to. Their volume (related to the 'shared_size' of the new class) is reflected by the fractional 'contribution' field. Since both stable_ids are listed in the primary key, they are not allowed to be NULLs. We shall treat empty strings as NULLs. If stable_id_from is empty, it means these members are newcomers into the new release. If stable_id_to is empty, it means these previously known members are disappearing in the new release. If both neither stable_id_from nor stable_id_to is empty, these members are truly migrating.
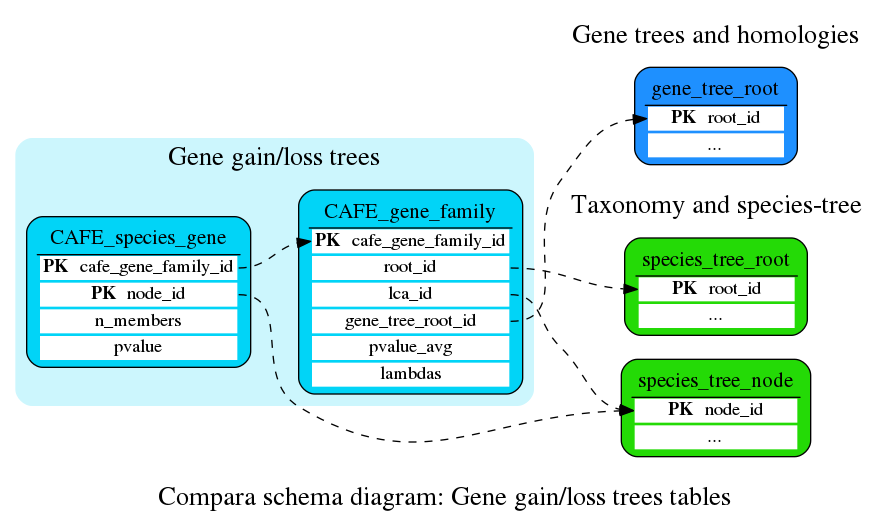
Gene gain/loss trees
Analysis of gain and loss across gene-families
This table holds information about each CAFE gene family
| See also: |
This table stores per species_tree_node information about expansions/contractions of each CAFE_gene_family
| See also: |
 Show columns
Show columns